Browse Articles
Conditioning Medicine
International bi-monthly journal of cell signaling, tissue protection, and translational research.
Current Location:Home / Browse Articles
Korean red ginseng mediates mitochondrial membrane potential repair via the Tom22-Tom20-SIRT2 pathway in astrocytes
Time:2022-10-15
Number:6172
Hui Su Jeon1, Chang-Hee Kim2, Minsu Kim1, Sunhong Moon1, Yoon Kyung Choi1
Author Affiliations


- 1Bio/Molecular Informatics Center, Department of Bioscience and Biotechnology, Konkuk University, Seoul 05029, Republic of Korean.
- 2Department of Otorhinolaryngology-Head and Neck Surgery, Konkuk University Medical Center, Konkuk University School of Medicine, Republic of Korea.
Conditioning Medicine 2022. 5(3): 105-111.
Abstract
Korean red ginseng extract (KRGE) may promote astrocytic mitochondrial function under pathophysiological conditions. The molecular mechanisms of KRGE-induced astrocytic mitochondrial activity involve nicotinamide phosphoribosyltransferase (Nampt), peroxisome proliferator-activated receptor-γ coactivator-1α (PGC-1α), and estrogen-related receptor α (ERRα) in oxygen-glucose deprivation/recovery (OGD/R)-conditioned astrocytes. In addition, the translocase of the outer membrane of mitochondria (Tom) 20 pathway in astrocytes is activated by KRGE. In this study, we investigated the relationship between Tom22 and Tom20, upstream signaling cascades (Nampt, PGC-1α, and ERRα) of the Tom22-Tom20 pathway, and the pairing effects of Tom20 and Tom22 on NAD+-dependent class I sirtuins (SIRTs), such as IRT2, in KRGE-treated astrocytes. We found that KRGE enhanced Tom22 and Tom20 expression via the induction of the Nampt-PGC-1α-ERRα circuit in astrocytes. SIRT2 may be imported into the mitochondria through the Tom22-Tom20 pathway. Consequently, SIRT2 may regulate oxidative phosphorylation complexes I and II, and mitochondrial membrane potential to mediate mitochondrial activity in astrocytes. Taken together, KRGE-mediated Tom22-Tom20-SIRT2 signaling may repair the OGD/R-mediated impairment of mitochondrial membrane potential and oxidative phosphorylation.
Keywords: Korean red ginseng extract (KRGE), The translocase of the outer membrane of mitochondria (Tom) 20, Tom22, Sirtuin 2 (SIRT2), Mitochondria membrane potential
Abstract
Korean red ginseng extract (KRGE) may promote astrocytic mitochondrial function under pathophysiological conditions. The molecular mechanisms of KRGE-induced astrocytic mitochondrial activity involve nicotinamide phosphoribosyltransferase (Nampt), peroxisome proliferator-activated receptor-γ coactivator-1α (PGC-1α), and estrogen-related receptor α (ERRα) in oxygen-glucose deprivation/recovery (OGD/R)-conditioned astrocytes. In addition, the translocase of the outer membrane of mitochondria (Tom) 20 pathway in astrocytes is activated by KRGE. In this study, we investigated the relationship between Tom22 and Tom20, upstream signaling cascades (Nampt, PGC-1α, and ERRα) of the Tom22-Tom20 pathway, and the pairing effects of Tom20 and Tom22 on NAD+-dependent class I sirtuins (SIRTs), such as IRT2, in KRGE-treated astrocytes. We found that KRGE enhanced Tom22 and Tom20 expression via the induction of the Nampt-PGC-1α-ERRα circuit in astrocytes. SIRT2 may be imported into the mitochondria through the Tom22-Tom20 pathway. Consequently, SIRT2 may regulate oxidative phosphorylation complexes I and II, and mitochondrial membrane potential to mediate mitochondrial activity in astrocytes. Taken together, KRGE-mediated Tom22-Tom20-SIRT2 signaling may repair the OGD/R-mediated impairment of mitochondrial membrane potential and oxidative phosphorylation.
Keywords: Korean red ginseng extract (KRGE), The translocase of the outer membrane of mitochondria (Tom) 20, Tom22, Sirtuin 2 (SIRT2), Mitochondria membrane potential
Introduction
Ischemia/reperfusion (I/R) injury in the central nervous system (CNS) triggers oxidative stress and mitochondrial dysfunction (Watson et al., 2013; O'Donnell et al., 2016). Astrocytes, one of the major glial cell types in the brain, actively modulate signaling and metabolism in the CNS by regulating mitochondrial function (Motori et al., 2013; Kim et al., 2019). The molecular mechanisms regulating astrocytic mitochondrial function involve peroxisome proliferator-activated receptor-γ coactivator-1α (PGC-1α) and estrogen-related receptor α (ERRα) (Choi et al., 2017; Kim et al., 2021a). Nicotinamide phosphoribosyltransferase (Nampt) catalyzes nicotinamide adenine dinucleotide (NAD) salvage synthesis in mammals (Wang and Miao, 2019). Sirtuins (silent information regulators, SIRT1–3), NAD+-dependent class I proteins, are upregulated by Nampt (Kim et al., 2022). Among SIRT1–7, Liu et al. (2017) found that SIRT2 directs and regulates mitochondrial metabolism. Furthermore, SIRT2 deficiency alters mitochondrial function by reducing striatal adenosine triphosphate (ATP) levels and increasing oxidative stress (Liu et al., 2017).
Translocases of the outer membrane of mitochondria (TOM complex) are multimeric proteins that serve as mitochondrial protein imports at the outer membrane (Anderson et al., 2019; Zhao and Zou, 2021). Tom40 forms the pore of the translocase, whereas Tom20, Tom22, and Tom70 function as receptors. Tom20 and Tom22 perform cooperatively through common signal recognition pathways as receptors for presequence-carrying precursors (Yamano et al., 2008).
Korean red ginseng extract (KRGE) promotes astrocytic mitochondrial function under pathophysiological conditions (Kim et al., 2021a; Kim et al., 2022). Although mitochondrial dysfunction is evident in neurodegenerative diseases (Goyal and Chaturvedi, 2021; O'Donnell et al., 2016; Watson et al., 2013), the underlying repair mechanisms remain unclear. Thus, in the present study, we dissected the signaling pathway involving Nampt, PGC-1α, ERRα, Tom20, Tom22, and SIRT2 in KRGE-mediated mitochondrial repair in astrocytes during oxygen-glucose deprivation (OGD) followed by recovery (OGD/R). We investigated the relationship between Tom22 and Tom20, the upstream signaling cascade involving Nampt, PGC-1α, and ERRα in the Tom22-Tom22 pathway, and the combinatory effects of Tom22 and Tom20 on NAD+-dependent class I SIRTs (i.e., SIRT2) in KRGE-treated astrocytes. We hypothesized that SIRT2 localizes to the mitochondria via the Tom22-Tom20 pathway, consequently upregulating mitochondrial membrane potential and oxidative phosphorylation via complex I (NDUFB8, NADH: ubiquinone oxidoreductase subunit B8) and complex II (SDHB, succinate dehydrogenase complex iron-sulfur subunit B).
Materials and methods
Materials
KRGE was obtained from the Korea Ginseng Corporation and stored at 4 °C. Stock solutions were prepared in filtering distilled states of water. The aliquoted stock solutions (200 mg/mL) were stored at -25 °C in the dark.
Cell culture and conditioned media preparation
Primary human brain astrocytes were acquired from the Applied Cell Biology Research Institute (Kirkland, WA, USA). Astrocytes were cultured in Dulbecco’s modified Eagle medium (DMEM, HyClone, Omaha, NE, USA) supplemented with 10% fetal bovine serum (FBS, Corning, NY, USA) and 1% penicillin/streptomycin (HyClone).
Oxygen glucose deprivation (OGD)/recovery (OGD/R)
After reaching 80% confluency, the primary human brain astrocytes were incubated with DMEM containing 0% FBS (no glucose, Thermo Fisher Scientific, Carlsbad, CA, USA) and 1% penicillin/streptomycin. The normoxic samples were incubated with 0% FBS-containing DMEM (with glucose, HyClone). Hypoxia was induced by perfusing 90% N2, 5% CO2, and 5% H2-containing gas for 15 min in a hypoxia chamber (Billups-Rothenberg, San Diego, CA, USA), and then placed for 8 h in a 37 °C incubator. After removing the dishes from the chamber, the media was replaced with DMEM containing 0% FBS and 1% penicillin/streptomycin and the cells were treated with the indicated concentrations of KRGE. The cells then underwent 24 h recovery at 37 °C in a 5% CO2 incubator under normoxic conditions (Thermo Fisher Scientific).
Transfection
At 70% confluency, the astrocytes were transfected with small interfering RNAs (siRNAs) against Nampt, SIRT2, Tom20, Tom22, PGC-1α, ERRα (50 nM, Santa Cruz Biotechnology, Dallas, TX, USA), or a negative control (50 nM, Thermo Fisher Scientific) using RNAiMax (Thermo Fisher Scientific). After approximately 14 h of recovery, cells were incubated under OGD conditions for 8 h and treated with or without KRGE for 24 h in serum-free DMEM.
Mitochondrial activity assay
Intracellular active mitochondria levels were assessed via quantitative fluorescence imaging using the mitochondrial membrane potential-sensitive dye (MitoTracker® Deep Red FM; Thermo Fisher Scientific). Astrocytes plated on 18-mm round coverslips in 12-well plates were cultured until they reached 70% confluency. Subsequently, cells were transfected with indicated siRNAs and were subjected to OGD/R with distilled water or 500 μg/mL of KRGE for 23.5 h. The cells were treated with MitoTracker (1 μM) for 30 min. After washing with phosphate-buffered saline (PBS), the cells were examined by immunocytochemical staining.
Mitochondria-enriched fraction
Cells were washed with PBS and collected using mannitol-sucrose buffer containing 210 mM mannitol (Sigma-Aldrich, Saint Louis, MO, USA), 70 mM sucrose (Sigma-Aldrich), 5 mM MOPS (Sigma-Aldrich), 1 mM EDTA (Sigma-Aldrich), and a protease inhibitor cocktail (Thermo Fisher Scientific), at pH 7.4. After the collected cells were homogenized with a 27-gauge 1/2-inch needle, the homogenates were centrifuged at 800 × g for 10 min to pellet the nuclei. The supernatants were used as total lysates or centrifuged at 10,000 × g for 20 min to collect a mitochondria-enriched fraction pellet. The pellets were then washed with a mannitol-sucrose buffer and centrifuged at 10,000 × g for 20 min. The resultant supernatants were filtered with a 3-kDa centrifugal filter (Merck, Darmstadt, Germany) at 4,000 × g for 2 h (cytosolic protein). The mitochondria-enriched fraction (pellet) was suspended in 30 μl Protein Extraction Solution (RIPA buffer) (Elpis-Biotech, Daejeon, South Korea), incubated for 20 min on ice, and centrifuged at 15,000 × g for 20 min. Ten μg mitochondrial protein (supernatant solution) and 10 μg cytosolic protein were then subjected to western blotting.
Immunocytochemistry
For immunocytochemistry, human astrocytes were seeded on coverslips in a 12-well incubating plate. After washing once with PBS to eliminate the media, the cells were fixed with 4% paraformaldehyde for 10 min. Cells were washed with PBST (0.1% Tween 20 [v/v] in PBS) for 10 min each. All subsequent washing steps were performed in the same manner. Cells were incubated with 3% bovine serum albumin (BSA) (US Biological Life Sciences, Salem, MA, USA) diluted in PBS containing 0.1% Triton X-100 (Sigma-Aldrich) for 1 h at room temperature to block non-specific binding. The primary antibodies were diluted in 0.3% BSA and added to each well. Samples were incubated with rabbit anti-SIRT2 (1:2000, Abcam, Cambridge, UK), mouse anti-Tom20 (1:200, Santa Cruz Biotechnology), and mouse anti-Tom22 (1:300, Santa Cruz Biotechnology) overnight at 4°C. After washing, the secondary antibody was diluted in 0.3% BSA and added to each well. Samples were incubated with a mixture of tetramethylrhodamine (TRITC)-conjugated donkey immunoglobulin G (IgG) (1:200; Jackson ImmunoResearch, West Grove, PA, USA) and fluorescein isothiocyanate (FITC)-conjugated donkey IgG (1:200; Jackson ImmunoResearch, West Frove, PA, USA) for 1 h at room temperature. Cells for MitoTracker detection were treated with FITC-conjugated donkey IgG. After the final wash, cells were mounted using Fluoro-Gel II with 4',6-diamidino-2-phenylindole (DAPI) mounting solution (Electron Microscopy Sciences, Hatfield, PA, USA) onto microslides. The upper side of the coverslips was reversed to allow attachment to the DAPI mounting solution. Images were acquired using an inverted microscope (Eclipse Ti2-U; Nikon, Tokyo, Japan).
Oxidative phosphorylation (OXPHOS) detection
Protein (15 μg) from the cell lysates were combined with sodium dodecyl sulfate (SDS) sample buffer (10% glycerol [v/v], Tris-Cl pH 6.8, 2% SDS [w/v], 1% β-mercaptoethanol [v/v], and bromophenol blue) and heated at 37 °C for 5 min. The protein samples were then subjected to western blot analysis. An OXPHOS antibody (1:5000; Abcam) was used. The OXPHOS antibody detects oxidative phosphorylation complex I (NDUFB8, NADH: ubiquinone oxidoreductase subunit B8), complex II (SDHB, succinate dehydrogenase complex iron-sulfur subunit B), complex III (UQCRC2, ubiquinol-cytochrome c reductase core protein 2), complex IV (MTCO1, cytochrome c oxidase subunit I), and complex V (ATP5A, ATP synthase).
Western blot analysis
RIPA buffer (Elpis-Biotech) was used for cell lysis. Selected amounts of protein from the cell lysates were combined with SDS sample buffer (10% glycerol [v/v], Tris-Cl pH 6.8, 2% SDS [w/v], 1% β-mercaptoethanol [v/v], and bromophenol blue) and incubated at 100 °C for 5 min. Protein samples were then separated using SDS polyacrylamide gel electrophoresis, transferred to polyvinylidene difluoride membranes (MilliporeSigma, Burlington, MA, USA), and blocked using Tris-buffered saline containing 0.1% Tween 20 and 5% skim milk (BD Difco, BD, Franklin Lakes, NJ, USA). The membranes were incubated with primary antibodies at 4 °C overnight. The primary antibodies used in this study were as follows: anti-Tom20 (1:1000, Santa Cruz Biotechnology; 1:3000, Abcam), anti-Tom22 (1:1000, Santa Cruz Biotechnology), anti-cytochrome c (Cyt c) (1:3000, BD Biosciences, San Jose, CA, USA), anti-Nampt (1:3000, AdipoGen Life Sciences, Liestal, Switzerland), anti-SIRT2 (1:3000, Abcam), anti-PGC-1α (1:1000, Santa Cruz Biotechnology), anti-ERRα (1:2000, Novus Biologicals, Littleton, CO, USA), and anti-β-actin (1:8000, Sigma-Aldrich). After washing, the membranes were incubated with peroxidase-conjugated secondary antibodies (1:8000, Thermo Fisher Scientific) and visualized using enhanced chemiluminescence (ECL; Elpis-Biotech) with appropriate western blot detection equipment (Fusion Solo, Vilber, Collégien, France).
Data analysis
Quantification of the intensity of protein expression from western blotting and immunocytochemistry experiments was performed using ImageJ (http://rsb.info.nih.gov/ij/, downloaded on 1 March 2022). GraphPad Prism 6 was used for statistical analyses. Multiple comparisons were evaluated using one-way analysis of variance (ANOVA) and Tukey’s post-hoc test (mean ± standard deviation). Statistical significance was set at p <0.05. *p < 0.05; **p < 0.01; ***p < 0.001.
Results
KRGE-induced Tom20 and Tom22 are restricted to mitochondria in astrocytes
To mimic I/R injury in an in vitro model, human astrocytes underwent OGD/R. Treatment of human astrocytes with KRGE during recovery significantly increased MitoTracker intensity compared to control (no KRGE) under OGD/R conditions (Fig. 1A–B). This effect was significantly blocked by the knockdown of Tom20 (si-Tom20) or Tom22 (si-Tom22) using specific siRNAs (Fig. 1A–B). Thus, KRGE-mediated changes in mitochondrial membrane potential may be regulated by the Tom20 and Tom22 pathways.
Since Tom20 immunoreactivity colocalized with MitoTracker staining (Fig. 1A), we isolated mitochondrial and cytosolic fractions. The expression of Tom20 and Tom22 in the mitochondrial fraction was increased by KRGE under OGD/R conditions compared with that of the non-KRGE treatment under OGD/R conditions (Fig. 1C). In this state, we did not detect any cytosolic expression of Tom20 or Tom22 (Fig. 1C). Next, we evaluated the relationship between Tom20 and Tom22 expression. KRGE-induced Tom20 was markedly reduced by Tom22 knockdown using si-Tom22 in astrocytes compared with that in non-KRGE treated astrocytes under OGD/R conditions (Fig. 1A, 1D), indicating that Tom22 acts as a strong Tom20 regulator. KRGE-enhanced Tom22 protein levels were slightly decreased by si-Tom20 (Fig. 1D), suggesting a cooperative interplay between Tom20 and Tom22.
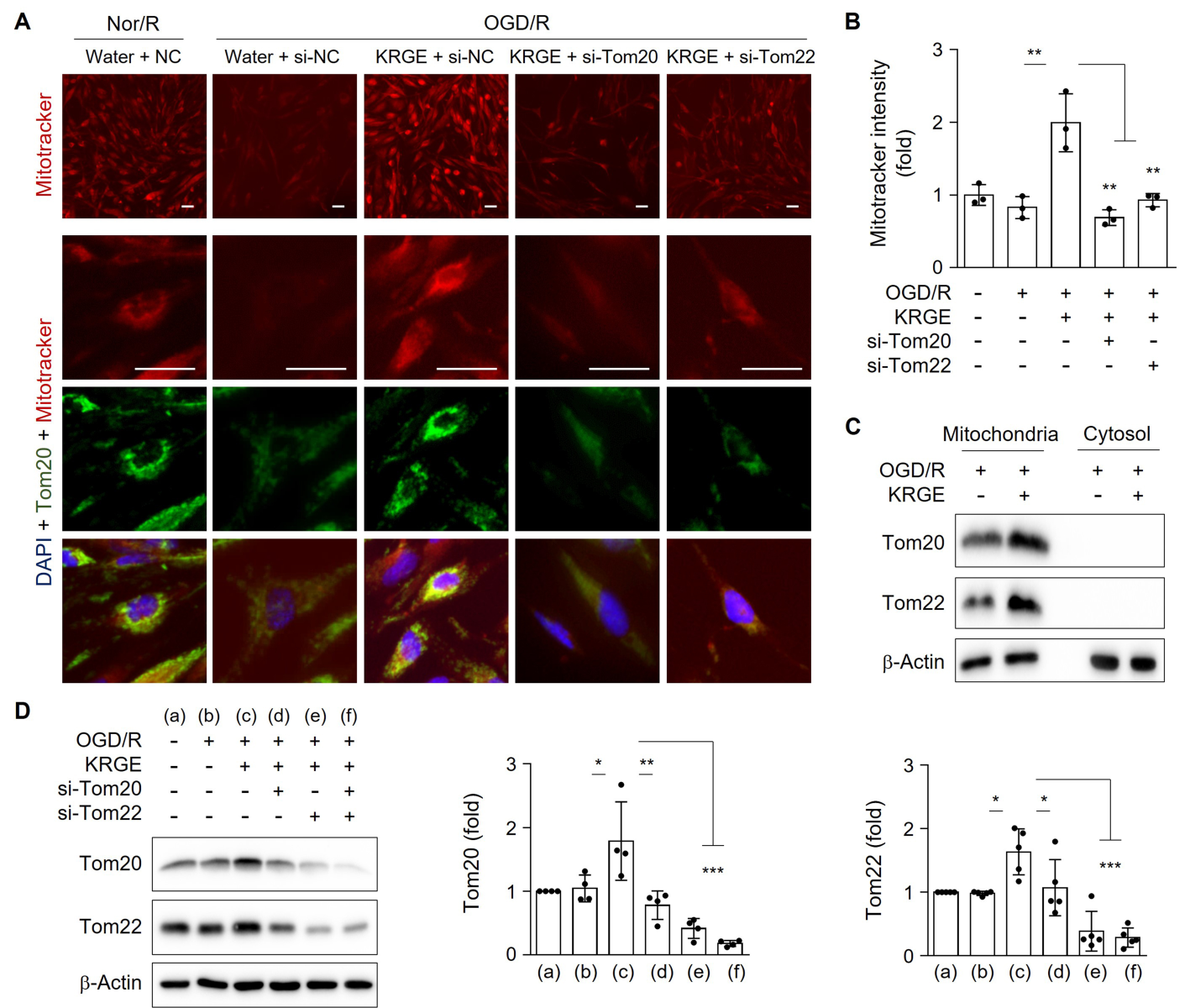
In a new window | Download PPT
Figure 1: KRGE-induced Tom20 and Tom22 are restricted to mitochondria in astrocytes. (A–B) Astrocytes on 12-well plates were transfected with si-Tom20 and/or si-Tom22 followed by OGD/R with 500 μg/mL KRGE. Normoxia followed by recovery (Nor/R) was used as a negative control for OGD/R. Representative image of human astrocytes stained with anti-Tom20 (green), MitoTracker (red), and DAPI (blue) (n = 3 per group) (A). Scale bar = 25 μm. Relative fluorescent intensity of randomized cells was measured using ImageJ (B). (C) Mitochondrial/cytosolic fractionation was performed. Protein levels were determined using western blotting (n = 3). (D) Human astrocytes were subjected to OGD/R with KRGE treatment for 24 h. Expression of the indicated proteins from human astrocytes was assessed by western blotting (n = 4–5 per group). Β-actin levels were used as an internal control. *p < 0.05; **p < 0.01; ***p < 0.001
KRGE induces Nampt-PGC-1α-ERRα complex-mediated Tom20 and Tom22 expression in astrocytes
Next, we examined the relationship between NAD+ synthesis-regulating proteins (i.e., Nampt) and mitochondrial function-related proteins (i.e., PGC-1α, ERRα, Tom20, and Tom22) in astrocytes. Astrocytes treated with KRGE under OGD/R conditions showed elevated expression of PGC-1α, ERRα, Tom20, and Tom22 relative to that in control cells (no KRGE treatment under OGD/R conditions), and these effects were effectively reduced by knockdown of Nampt using si-Nampt (Fig. 2A). The expression of the NAD+-dependent deacetylase SIRT2 was also regulated by Nampt (Fig. 2A). KRGE-mediated Nampt induction was diminished by si-ERRα (Fig. 2B), suggesting a positive feedback loop between Nampt and ERRα in KRGE-treated astrocytes. Interestingly, transfection of cells with both si-PGC-1α and si-ERRα siRNAs in KRGE-treated astrocytes under OGD/R conditions decreased the protein levels of Nampt, Tom20, Tom22, and SIRT2 compared with those in the non-KRGE treated astrocytes under OGD/R conditions (Fig. 2B). These data suggest that the Nampt-PGC-1α-ERRα signaling complex regulates Tom20, Tom22, and SIRT2 expression.
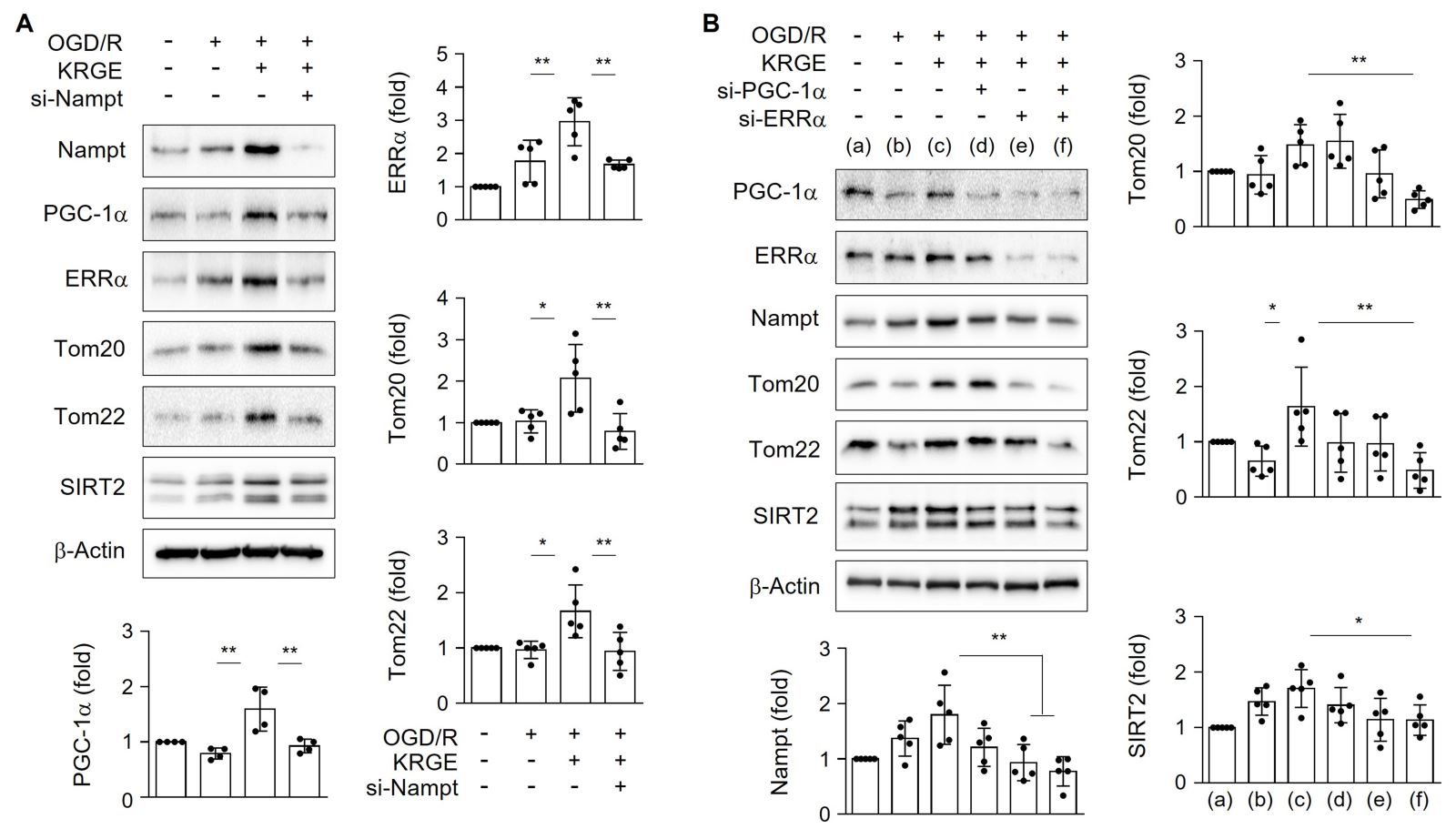
In a new window | Download PPT
Figure 2: KRGE induces Nampt-PGC-1α-ERRα complex-mediated Tom20 and Tom22 expression in astrocytes. (A) Human astrocytes were transfected with si-Nampt, and then subjected to OGD/R with 250 μg/mL KRGE. Indicated protein levels from lysed cells were detected by western blotting (n = 4–5 per group). (B) Human astrocytes were transfected with si-PGC-1α and/or si-ERRα, and then subjected to OGD/R with 250 μg/mL KRGE. Cells were lysed following various siRNA transfections. Indicated protein levels were detected by western blotting (n = 5 per group). *p < 0.05; **p < 0.01.
KRGE induces SIRT2 via the Tom22-Tom20 pathway
We next examined the roles of Tom20 and Tom22 in modulating mitochondrial function-regulating proteins in astrocytes exposed to KRGE under OGD/R conditions. Treatment of human astrocytes with both si-Tom20 and si-Tom22 significantly reduced the protein levels of OXPHOS complexes I and II, Cyt c, and SIRT2 after KRGE treatment compared with that in controls (non-KRGE treated astrocytes under OGD/R conditions) (Fig. 3A). SIRT2 immunoreactivity was increased in Tom22-expressing organelles, including the mitochondria, in KRGE-exposed astrocytes under OGD/R conditions compared with that in controls (Fig. 3B), which was reversed by the paired reduction of Tom20 and Tom22 (Fig. 3C). Taken together, the astrocyte-derived Tom22-Tom20 pathway may play a key role in regulating mitochondrial activity and SIRT2 expression in mitochondria under KRGE-treated OGD/R conditions.
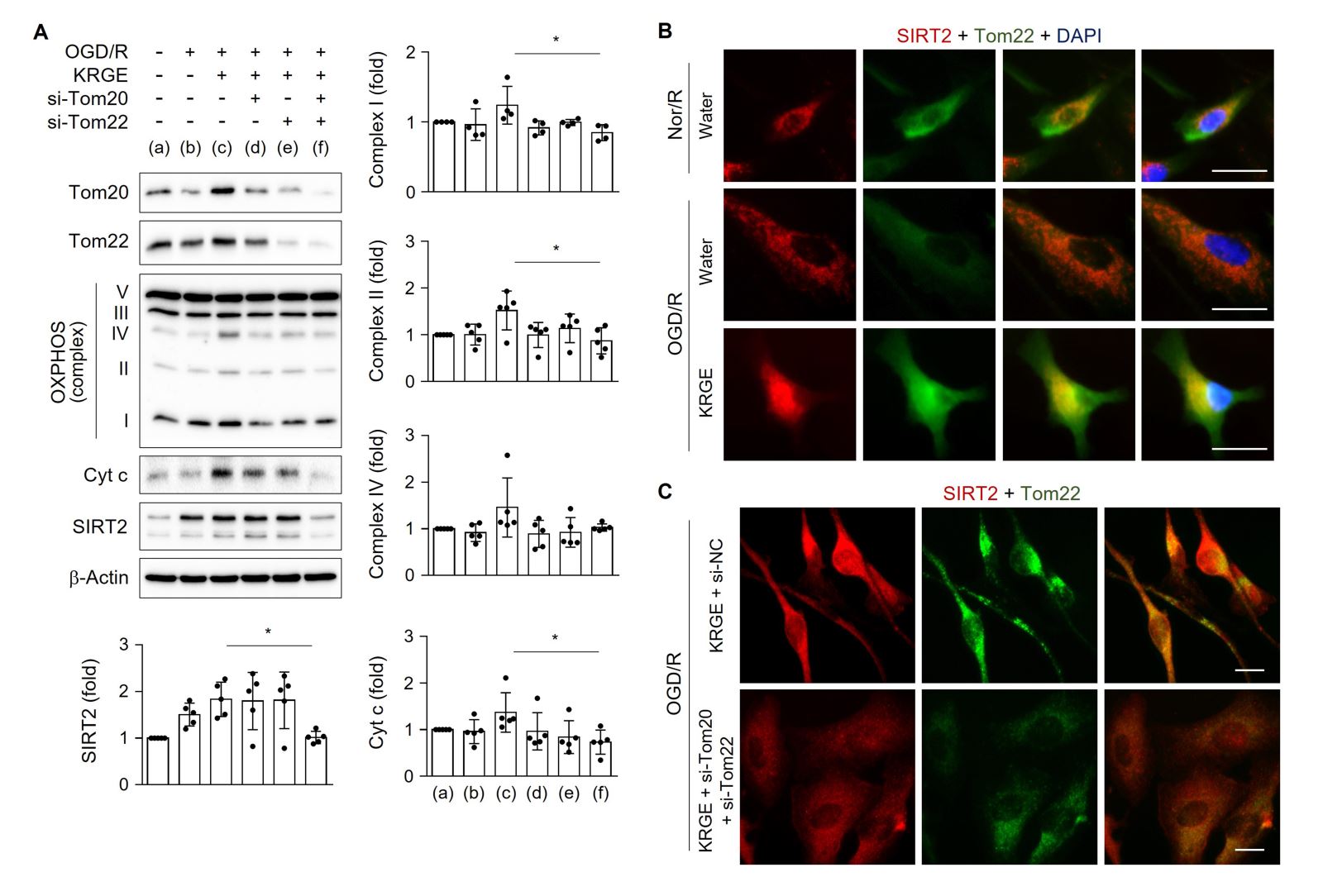
In a new window | Download PPT
Figure 3: KRGE induces SIRT2 via Tom20/Tom22. (A) Human astrocytes were transfected with si-Tom20 and/or si-Tom22, and then subjected to OGD/R with 250 μg/mL KRGE. Indicated protein levels from lysed cells were detected by western blotting (n = 4–5 per group). *p < 0.05. (B–C) Astrocytes on 12-well plates underwent OGD/R and were incubated with 500 μg/mL KRGE. Normoxia followed by recovery (Nor/R) was used as a control for OGD/R (B). Representative image of the anti-SIRT2 (red), anti-Tom22 (green), and DAPI (blue) staining in human astrocytes (n = 5 per group). Scale bar = 25 μm. (C) Astrocytes on 12-well plates were transfected with si-Tom20 and/or si-Tom22 followed by OGD/R with KRGE. Representative image of the anti-SIRT2 (red), anti-Tom22 (green), and DAPI (blue) staining in human astrocytes (n = 5 per group). Scale bar = 25 μm. The relative fluorescent intensity of randomized cells was measured using ImageJ (B–C).
KRGE induces SIRT2-mediated mitochondrial membrane potential in astrocytes
To examine the role of SIRT2 in mitochondrial function, astrocytes were transfected with si-SIRT2. MitoTracker intensity was significantly reduced by si-SIRT2 in KRGE-treated OGD/R cells compared with that of controls (Fig. 4A). Knockdown of SIRT2 was confirmed by immunocytochemistry (Fig. 4B). SIRT2 knockdown significantly decreased KRGE-mediated levels of OXPHOS complexes I and II, and Cyt c compared to that of controls (Fig. 4C). Collectively, these data suggest that SIRT2 acts as a key mediator of KRGE-induced mitochondrial membrane potential and oxidative phosphorylation under OGD/R conditions.
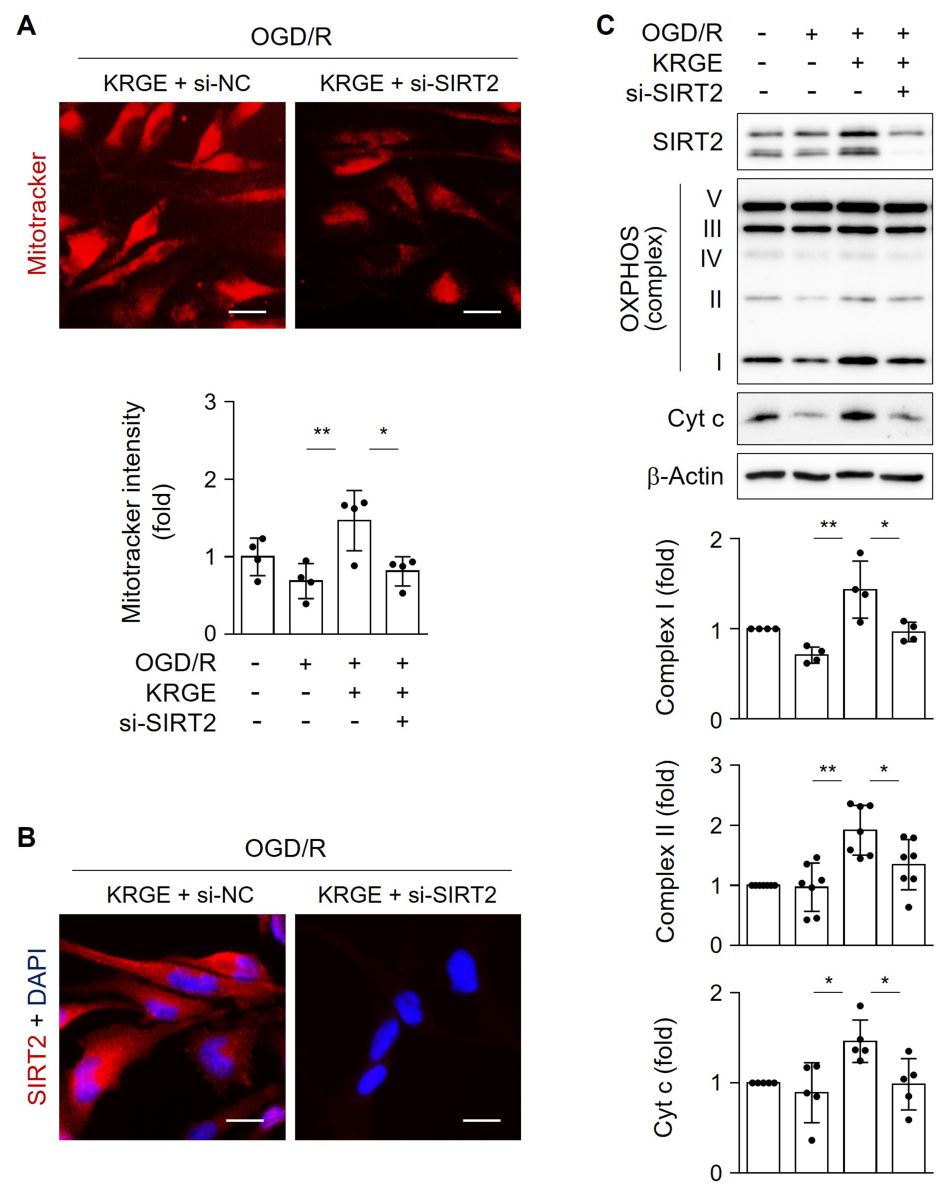
In a new window | Download PPT
Figure 4: KRGE induces SIRT2-mediated mitochondrial membrane potential in astrocytes. (A–B) Astrocytes on 12-well plates were transfected with si-control or si-SIRT2 followed by OGD/R with 500 μg/mL KRGE. (A) Representative image of the MitoTracker (red) staining in human astrocytes (n = 4 per group). Scale bar = 25 μm. (B) Representative image of the anti-SIRT2 (red) and DAPI (blue) staining in human astrocytes (n = 4 per group). Scale bar = 25 μm. (C) Astrocytes were transfected with si-SIRT2 followed by OGD/R with 250 μg/mL KRGE. Indicated protein expression was assessed by western blotting (n = 4–7 independent experiments). *p < 0.05; **p < 0.01.
Discussion
During I/R injury, damaged vascular cells may cause O2 and glucose deficiencies in the brain (Choi et al., 2016; Fraisl et al., 2009). KRGE has beneficial effects on I/R injury in the CNS (Kim et al., 2021b). Here, KRGE contain several ginsenosides such as ginsenoside Rb1 (5.85 mg/g), Rg3s (4.43 mg/g), Rc (2.29 mg/g), Rb2 (2.17 mg/g), Rg3r (2.02 mg/g), Rf (1.37 mg/g), and Rh1 (1.26 mg/g). Among them, ginsenoside Rc can activate SIRT1 and increase the levels of OXPHOS II-IV in cardiomyocytes and neuronal cells in normal and I/R injury conditions (Huang et al., 2021). However, the molecular mechanisms by which KRGE induces astrocytic mitochondrial function during I/R injury are not well established.
Our previous study showed that Nampt-induced intracellular NAD+ levels affect mitochondrial function through class I SIRTs (Kim et al., 2022). In this study, we found that KRGE induced a Nampt-ERRα circuit in astrocytes. Furthermore, the combination of PGC-1α and ERRα regulated Nampt, Tom22, Tom20, and SIRT2 more significantly than ERRα alone. The TOM complex located on the outer membrane of the mitochondria triggers protein import within organelles (Zhao and Zou, 2021). One of the TOM complexes, Tom22, restores levels of oxidative phosphorylation proteins, such as NADH dehydrogenase Fe-S protein 1 (NDUFS1, OXPHOS complex I) and SDHB (OXPHOS complex II), which are downregulated in high glucose-treated human umbilical vein endothelial cells (Zeng et al., 2019).
Translocation of SIRT2 to the inner mitochondrial membrane may relate to mitophagy (Liu et al., 2017). Impaired translocation of proteins into the mitochondria contributes to the pathogenesis of neurodegenerative diseases (Goyal and Chaturvedi, 2021). In this study, we found that KRGE-induced Tom22 and Tom20 may be involved in mitochondrial SIRT2 import and SIRT2-mediated oxidative phosphorylation of proteins such as complex I and II. Tom20 paired with Tom22 also regulated the expression of OXPHOS complex I and II, and Cyt c in KRGE-treated astrocytes.
In conclusion, the Nampt-PGC-1α-ERRα signaling complex regulates the expression of Tom22 and Tom20, which may be partly responsible for SIRT2 translocation in mitochondria. Tom22 regulates Tom20 expression and vice versa. These consequences may result in an increase in OXPHOS complexes I and II, as well as in mitochondrial membrane potential. As a putative therapeutic agent, KRGE may enhance the repair processes for I/R injury by boosting astrocytic mitochondrial activity through the Tom22-Tom20-SIRT2 pathway.
Author contributions
Jeon HS, Kim CH, and Choi YK designed the project and wrote the manuscript. Choi YK revised the manuscript accordingly. Jeon HS, Kim CH, Kim M, Moon S, and Choi YK performed the experiments and analyzed the data. Kim CH, Choi YK received funding for this study. Choi YK supervised the project.
Acknowledgements and sources of funding
This research was supported by a grant from the National Research Foundation of Korea (NRF) funded by the Korean Government (MSIP) (2020R1A2C1004397 & 2021R1F1A1062019). This research was also funded by a grant from the Korean Society of Ginseng (2021).
Conflict of interest
The authors declare that they have no conflicts of interest.
References
Hui Su Jeon1,†
1Bio/Molecular Informatics Center, Department of Bioscience and Biotechnology, Konkuk University, Seoul 05029, Republic of Korean.
Chang-Hee Kim2,†
2Department of Otorhinolaryngology-Head and Neck Surgery, Konkuk University Medical Center, Konkuk University School of Medicine, Republic of Korea.
Minsu Kim1
1Bio/Molecular Informatics Center, Department of Bioscience and Biotechnology, Konkuk University, Seoul 05029, Republic of Korean.
Sunhong Moon1
1Bio/Molecular Informatics Center, Department of Bioscience and Biotechnology, Konkuk University, Seoul 05029, Republic of Korean.
Yoon Kyung Choi1*
1Bio/Molecular Informatics Center, Department of Bioscience and Biotechnology, Konkuk University, Seoul 05029, Republic of Korean.
Hui Su Jeon and Chang-Hee Kim contributed equally to this article.
Corresponding author:
Yoon Kyung Choi, PhD
Email: ykchoi@konkuk.ac.kr

In a new window | Download PPT
Figure 1: KRGE-induced Tom20 and Tom22 are restricted to mitochondria in astrocytes. (A–B) Astrocytes on 12-well plates were transfected with si-Tom20 and/or si-Tom22 followed by OGD/R with 500 μg/mL KRGE. Normoxia followed by recovery (Nor/R) was used as a negative control for OGD/R. Representative image of human astrocytes stained with anti-Tom20 (green), MitoTracker (red), and DAPI (blue) (n = 3 per group) (A). Scale bar = 25 μm. Relative fluorescent intensity of randomized cells was measured using ImageJ (B). (C) Mitochondrial/cytosolic fractionation was performed. Protein levels were determined using western blotting (n = 3). (D) Human astrocytes were subjected to OGD/R with KRGE treatment for 24 h. Expression of the indicated proteins from human astrocytes was assessed by western blotting (n = 4–5 per group). Β-actin levels were used as an internal control. *p < 0.05; **p < 0.01; ***p < 0.001

In a new window | Download PPT
Figure 2: KRGE induces Nampt-PGC-1α-ERRα complex-mediated Tom20 and Tom22 expression in astrocytes. (A) Human astrocytes were transfected with si-Nampt, and then subjected to OGD/R with 250 μg/mL KRGE. Indicated protein levels from lysed cells were detected by western blotting (n = 4–5 per group). (B) Human astrocytes were transfected with si-PGC-1α and/or si-ERRα, and then subjected to OGD/R with 250 μg/mL KRGE. Cells were lysed following various siRNA transfections. Indicated protein levels were detected by western blotting (n = 5 per group). *p < 0.05; **p < 0.01.

In a new window | Download PPT
Figure 3: KRGE induces SIRT2 via Tom20/Tom22. (A) Human astrocytes were transfected with si-Tom20 and/or si-Tom22, and then subjected to OGD/R with 250 μg/mL KRGE. Indicated protein levels from lysed cells were detected by western blotting (n = 4–5 per group). *p < 0.05. (B–C) Astrocytes on 12-well plates underwent OGD/R and were incubated with 500 μg/mL KRGE. Normoxia followed by recovery (Nor/R) was used as a control for OGD/R (B). Representative image of the anti-SIRT2 (red), anti-Tom22 (green), and DAPI (blue) staining in human astrocytes (n = 5 per group). Scale bar = 25 μm. (C) Astrocytes on 12-well plates were transfected with si-Tom20 and/or si-Tom22 followed by OGD/R with KRGE. Representative image of the anti-SIRT2 (red), anti-Tom22 (green), and DAPI (blue) staining in human astrocytes (n = 5 per group). Scale bar = 25 μm. The relative fluorescent intensity of randomized cells was measured using ImageJ (B–C).

In a new window | Download PPT
Figure 4: KRGE induces SIRT2-mediated mitochondrial membrane potential in astrocytes. (A–B) Astrocytes on 12-well plates were transfected with si-control or si-SIRT2 followed by OGD/R with 500 μg/mL KRGE. (A) Representative image of the MitoTracker (red) staining in human astrocytes (n = 4 per group). Scale bar = 25 μm. (B) Representative image of the anti-SIRT2 (red) and DAPI (blue) staining in human astrocytes (n = 4 per group). Scale bar = 25 μm. (C) Astrocytes were transfected with si-SIRT2 followed by OGD/R with 250 μg/mL KRGE. Indicated protein expression was assessed by western blotting (n = 4–7 independent experiments). *p < 0.05; **p < 0.01.
Supporting Information
Metrics
| Full-Text | Supporting Information | ||
|---|---|---|---|
| Number | 6172 | 15 | 0 |
Copyright © 2017 Conditioning Medicine, All Rights Reserved.
Address: Conditioning Medicine Editorial Office, 3500 Terrace Street, Pittsburgh, PA, 15213, USA